
Peking University, October 31, 2024: On October 23, research teams headed by Peking University Department of Computer Science Dean Cheng Zhang and Peking University Center for Quantitative Biology researcher Long Qian published their article in Nature titled “Parallel molecular data storage by printing epigenetic bits on DNA.”
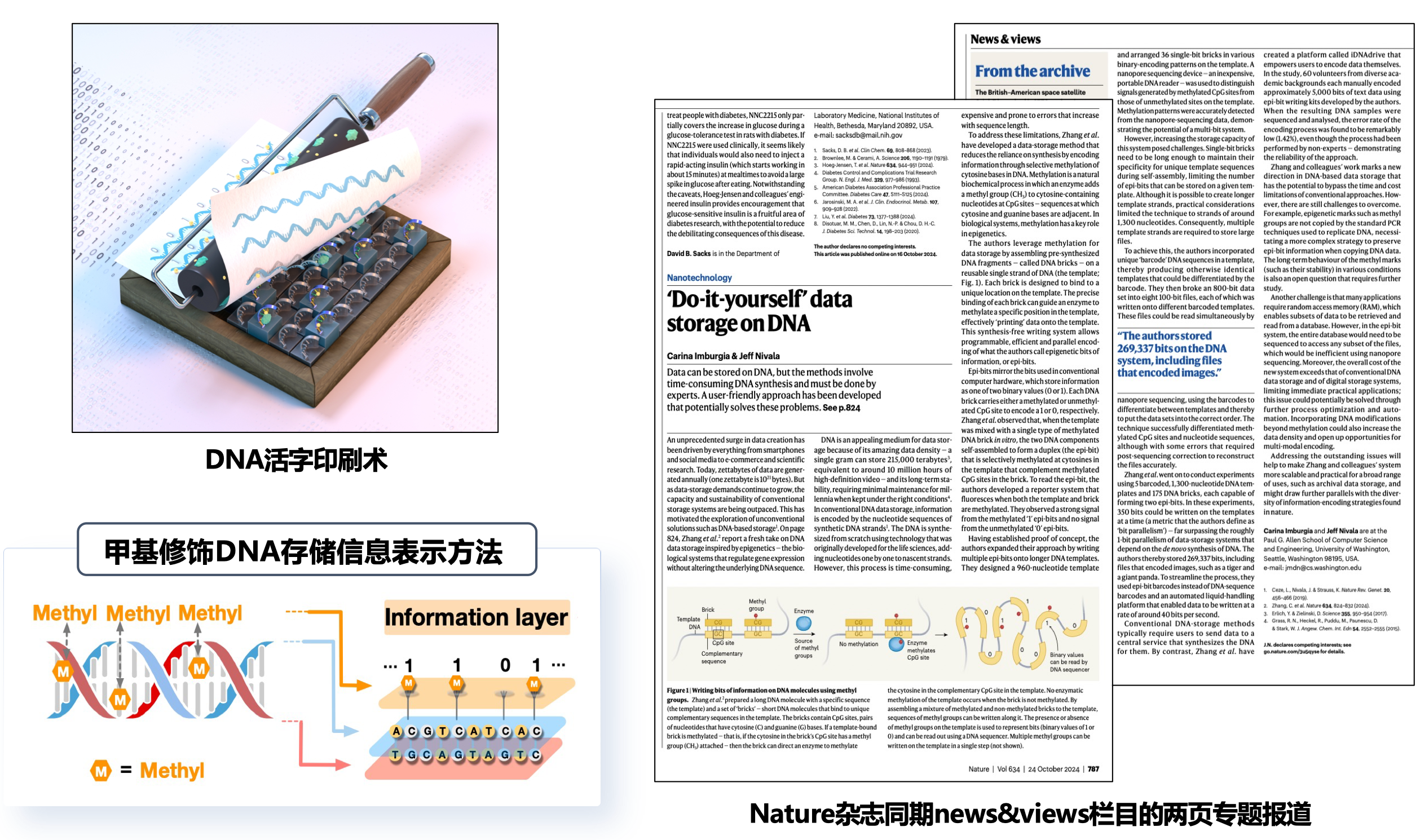
Their research introduces a novel method for DNA storage. Through enzymatic methylation, bits of data in the form of epigenetic modifications (“epi-bits”) can be precisely introduced onto universal DNA templates.
Why it matters:
In the era of big data, global mass data flow has presented data storage systems with a looming challenge. As DNA has incredibly high storage density – a single gram of DNA can store 215,000 terabytes, the same size as 10 million hours of high-definition video (Imburgia & Nivala, 2024)– and long-term stability, it is an attractive medium for data storage. However, conventional DNA storage relies on de novo synthesis, where nucleotides are added one by one in a fixed order, making the process time-consuming and costly. Zhang et al.’s method enables DNA self-assembly that allows data writing to become parallel and programmable.
Additionally, the epi-bit method can be used by individuals to personalize their DNA storage, as shown by implementation of the method by 60 volunteers from diverse academic backgrounds. This clearly demonstrates the potential Zhang et al.’s epi-bit method has as an accessible, versatile, fast, and low-cost method for DNA storage.
Methodology:
- Information is encoded through selective methylation on cytosine bases in DNA.
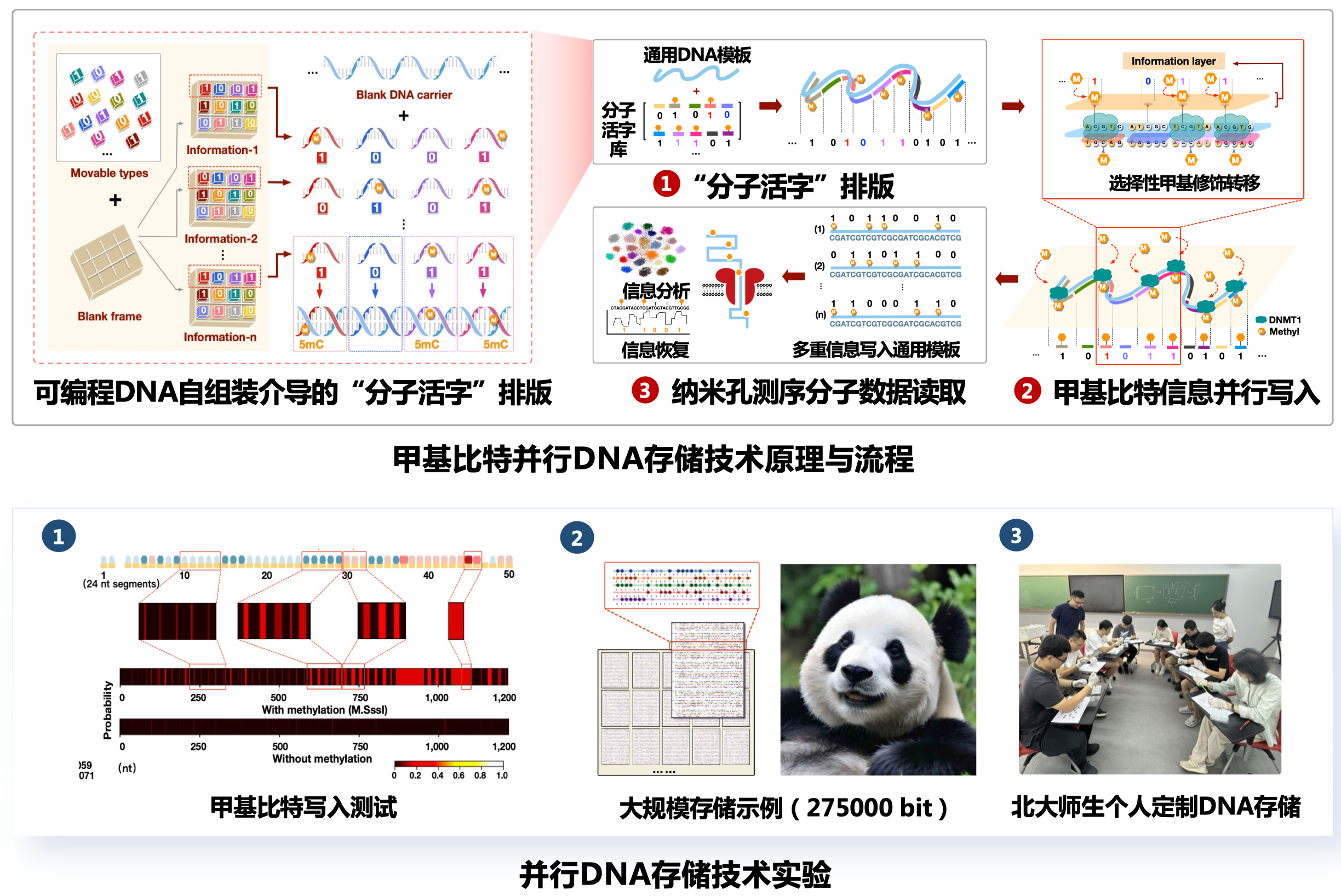
- Pre-synthesized DNA fragments, called DNA bricks, are assembled on a reusable DNA strand. Each DNA brick binds to a unique location on the strand.
- The precise binding of the brick guides an enzyme to methylate a specific position on the template, which effectively “prints” the data onto the template.
- Following the same binary system as computer hardware, each DNA brick carries a methylated or unmethylated site to encode a 1 or 0, respectively.
- Epi-bits are read using a nanopore sequencing device.
Key findings:
- Using the epi-bit method, Zhang et al. were able to write 275,000 information bits onto five templates on an automated platform, with no synthesizing of DNA required, including two high-definition photos of a white tiger and a giant panda.
- On iDNAdrive, a platform created by Zhang et al. that allows users to encode data themselves, volunteers encoded approximately 5,000 bits of data using epi-bit writing kits. The error rate when reading the data was as low as 1.42%.
Written by: Zhou Runxi
Edited by: Wang Mengjiao
Source: PKU News (Chinese)


