Peking University, Oct. 16, 2018: Tang Fuchou research team from Peking University Beijing Advanced Innovation Center for Genomics and School of Life Sciences Biomedical Pioneering Innovation Center and Qiao Jie research team from PKU Third Hospital jointly published an online paper in Cell Reports titled “Dissecting the Global Dynamic Molecular Profiles of Human Fetal Kidney Development by Single-Cell RNA Sequencing.” The research applied single-cell RNA sequencing to more than 3000 renal cells spanning several embryonic stages from 7 to 25 weeks. Centering on nephron formation during the kidney development, the research details the heterogeneous composition of nephron progenitors, analyses the transcription regulation and signaling pathway mechanism in the process of nephron progenitors segmenting into different nephron epithelial tubules. The research also highlights the expression features of causal genes of congenital renal diseases during fetal kidney development.
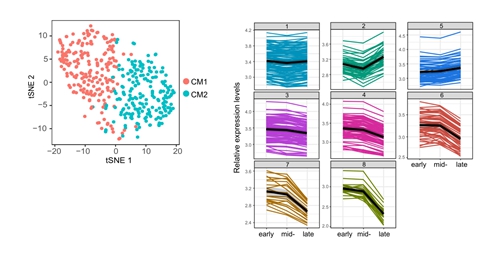
Figure 1. Molecular Features of Cap Mesenchyme
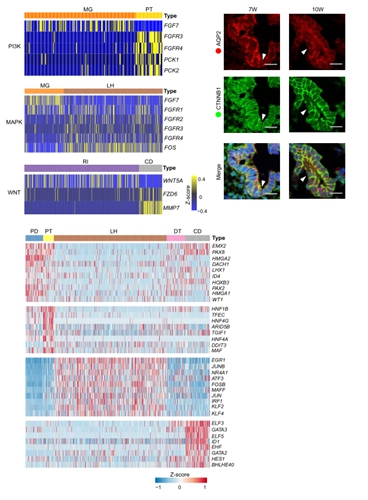
Figure 2. Regulators and Signaling Pathways Involved in Nephron Tubule Segmentation

Figure 3. Expression Tendencies of AQP2 and CA2 in the Development of Collecting Duct Epithelium

Figure 4. Expression Features of Risk Caudal Genes of Different Congenital Renal Diseases
The study is of great significance in terms of having a better understanding of human kidney development and treating congenital renal diseases. Meanwhile, the research profiles the regulatory elements present in heterogeneous renal cell development, which will facilitate new approaches to renal cell induction in vitro and disease treatment.
Written by: Lang Lang
Edited by: Zhang Jiang
Source: PKUHSC