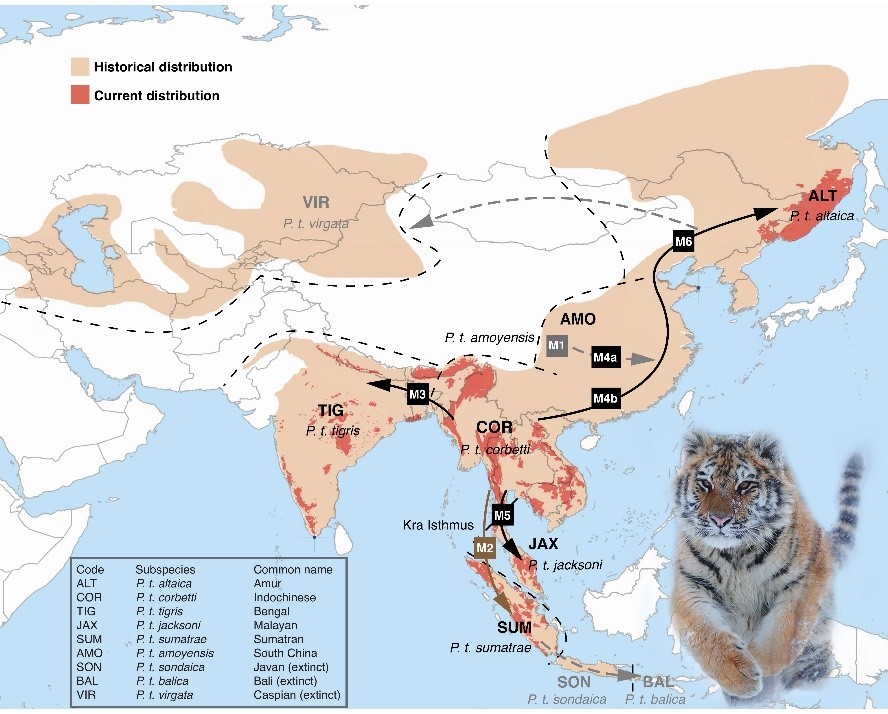
Peking University, Oct. 26, 2018: Fewer than 4,000 free-ranging tigers remain in the wild. Efforts to protect these remaining tigers have also been stymied by uncertainty about whether they represent six, five or only two subspecies. Now, researchers who've analyzed the complete genomes of 32 representative tiger specimens confirm that tigers indeed fall into six genetically distinct groups. The findings are reported in Current Biology on October 25. These six subspecies include the Bengal tiger, Amur tiger, South China tiger, Sumatran tiger, Indochinese tiger, and Malayan tiger, first proposed in 2004. Three other tiger subspecies have already been lost to extinction.
"The lack of consensus over the number of tiger subspecies has partially hindered the global effort to recover the species from the brink of extinction, as both captive breeding and landscape intervention of wild populations increasingly requires an explicit delineation of the conservation management units," says Luo Shu-Jin of Peking University in Beijing. "This study is the first to reveal the tiger's natural history from a whole-genomic perspective. It provides robust, genome-wide evidence for the origin and evolution of this charismatic megafauna species."
Luo and colleagues, including first author Liu Yue-Chen, set out to expand their earlier genetic evidence on tiger's evolutionary history and population structure using a whole-genome approach. They realized that genome-wide screening was also the only way to look for signals that distinct groups of tigers have undergone natural selection to adapt to the environments found in the distinct geographic regions they inhabit.
Fossil evidence shows that tigers go back two to three million years. But, the genomic evidence shows that all living tigers only trace back to a time about 110,000 years ago, when tigers suffered a historic population bottleneck. The genomic evidence shows that there is very little gene flow among tiger populations. Despite the tiger's low genetic diversity, the pattern across groups is highly structured, offering evidence that these subspecies each have a unique evolutionary history. That's quite unique among the big cats, the researchers say, noting that several other species, such as the jaguar, have shown much more evidence of intermixing across whole continents.
Tiger subspecies have distinct features. For example, Amur tigers are large with pale orange fur, while Sumatran tigers in the Sunda Islands tend to be smaller with darker, thickly striped fur. In fact, despite the very recent common ancestor of all living tigers, the researchers were able to detect evidence of natural selection.
"In the end, we were quite amazed that, by performing a stepwise genome-wide scan, seven regions including 14 genes stood out as the potential regions subject for selection," Luo says.
The strongest signal of selection they found was in the Sumatran tiger, across a genomic region that contains the body-size-related ADH7 gene. The researchers suggest that the Sumatran tiger might have been selected for smaller size to reduce its energy demands, allowing it to survive on the island's smaller prey animals, such as wild pigs and muntjac, a small deer.
The new findings provide the strongest genetic evidence yet for subspecies delineation in tigers. "Tigers are not all alike," Luo says. "Tigers from Russia are evolutionarily distinct from those from India. Even tigers from Malaysia and Indonesia are different."
The origin of the South China tiger remains unresolved since only one specimen from captivity was used in this study, the researchers note. Unfortunately, this subspecies has gone extinct in the wild. The researchers plan to study old specimens with known origin from all over China to fill in the missing pieces of living tigers' evolutionary history. They're also retrieving genomic information from historical specimens, including those representing the extinct Caspian, Javan, and Bali tigers.
This map shows postulated dispersal routes and range expansions of modern tigers.
Edited by: Qiu Tianjie
Source: School of Life Sciences